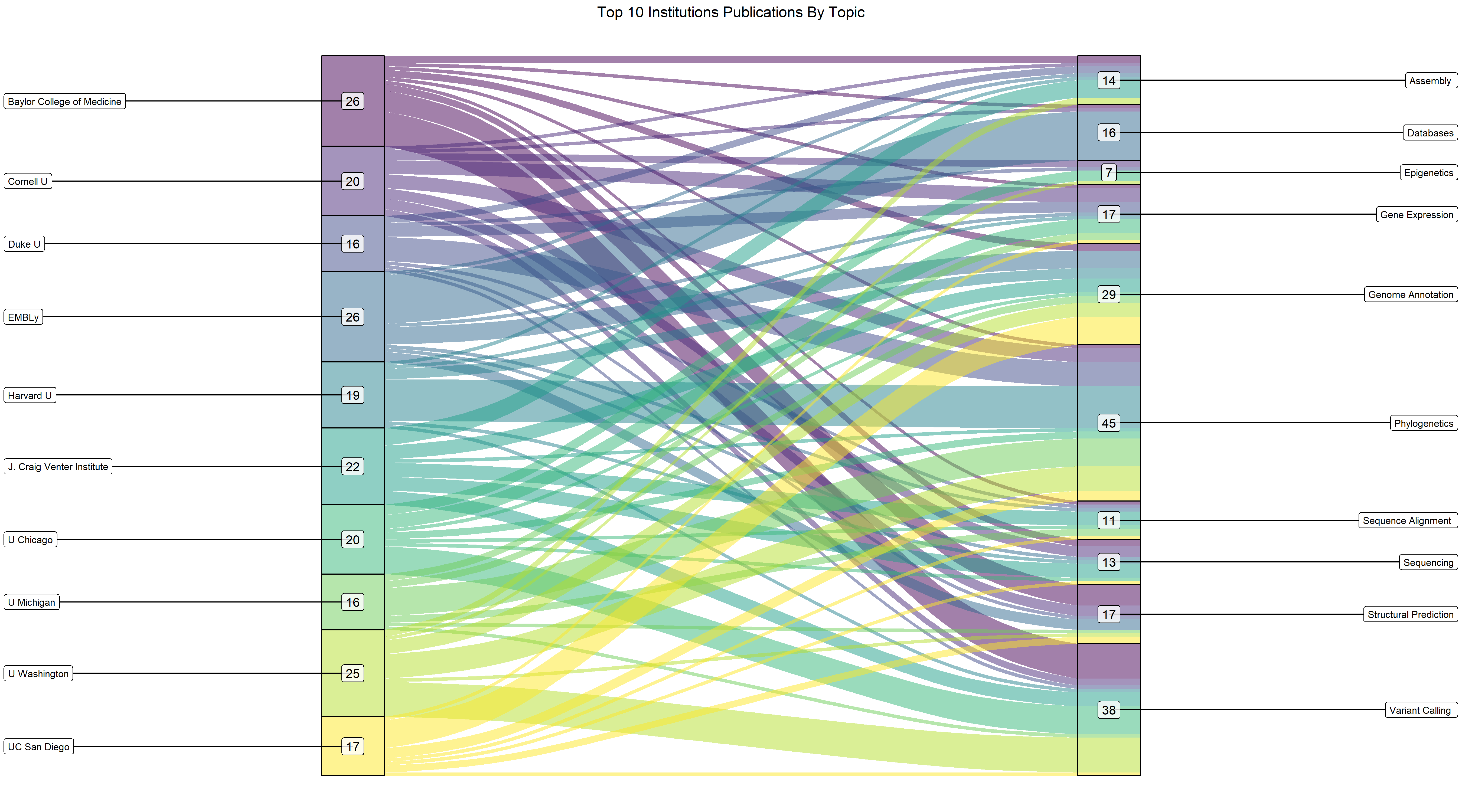
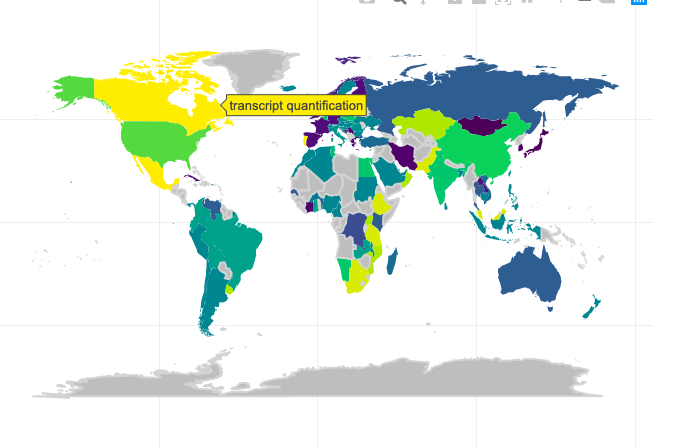
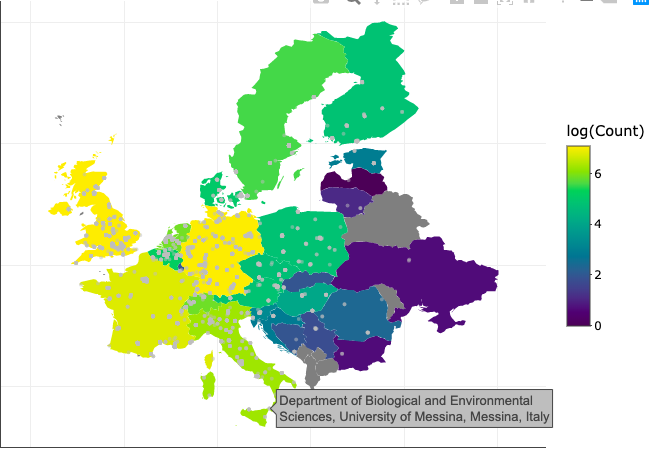
Here’s an overview of plotting the Sankey diagram using ggplot:
sankey <- ggplot(datSK, aes(y = Weight, axis1 = From, axis2 = To)) +
geom_alluvium(aes(fill = From), width = 1 / 12) +
geom_stratum(alpha = 0, width = 1 / 12, color = "black") +
scale_x_discrete(limits = c("From", "To"), expand = c(0.3, 0.1)) +
scale_fill_viridis_d() +
theme_void() +
theme(axis.title.y = element_blank(), axis.title.x = element_blank(),
axis.ticks.x = element_blank(), axis.ticks.y = element_blank(),
axis.text.x = element_blank(),axis.text.y = element_blank(),
legend.position = "none", plot.title = element_text(hjust = 0.5)) +
ggrepel::geom_label_repel(
aes(label = From), stat = "stratum", size = 3, direction = "x", hjust = 10) +
ggrepel::geom_label_repel(
aes(label = To), stat = "stratum", size = 3, direction = "y", nudge_x = 0.5) +
geom_label(aes(label = Weight), stat = "stratum", alpha = 0.8) +
ggtitle("Top 10 Institutions Publications By Topic")
sankey <- set_panel_size(sankey, width = unit(18, "cm"), height = unit(10, "cm"))
grid.newpage()
grid.draw(sankey)